by Microbiology Doctor-dr
Primer- Definition, Types, Primer Design Online Tools, Uses
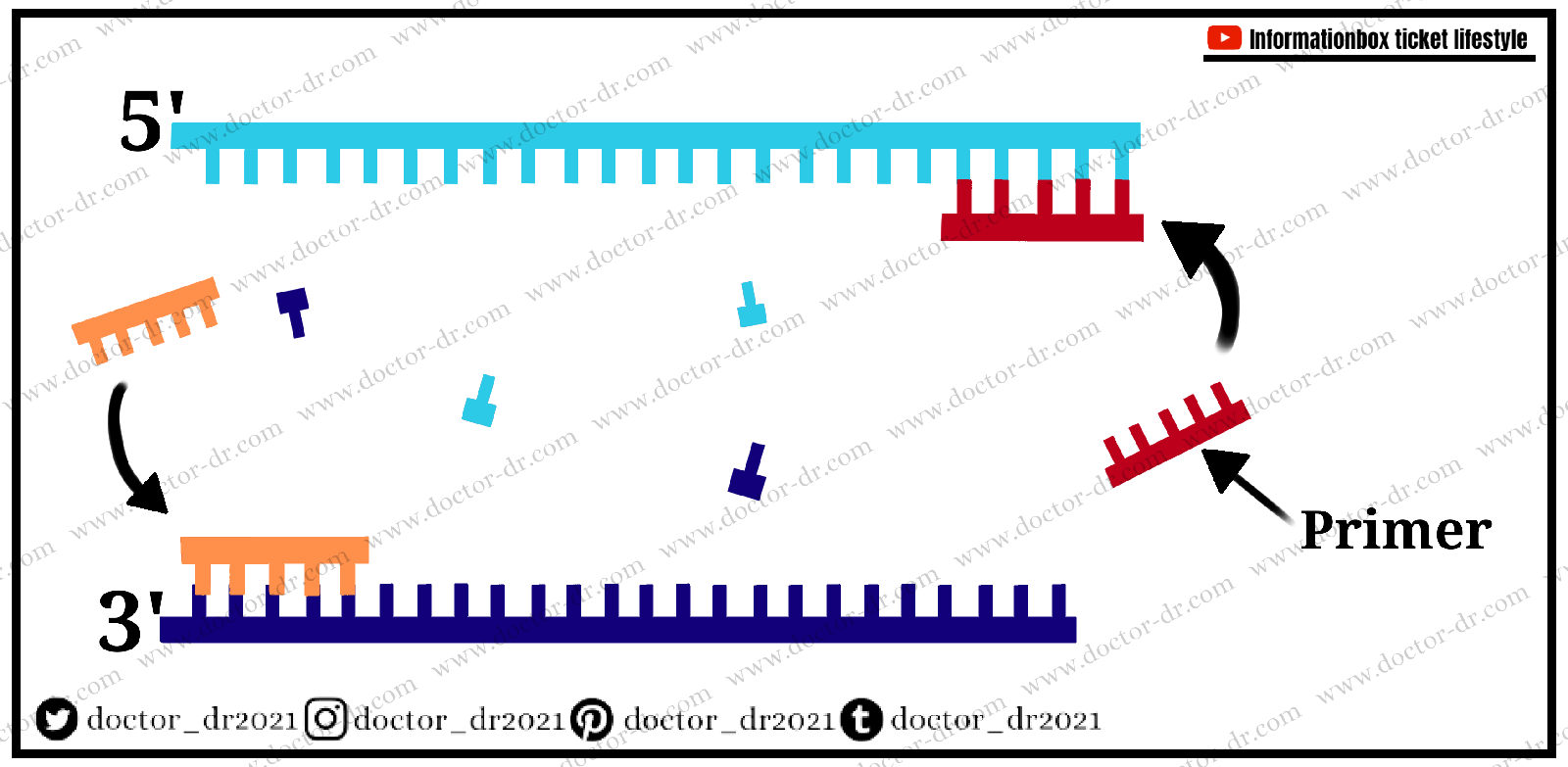
What is Primer?
The idea of stability in DNA primers as opposed to RNA primers.
A primer is a brief sequence that acts as the starting point for DNA synthesis. A point of initiation synthesis can be a set of primers (forward and reverse) having a sequence complementary to the template DNA.
The primer's primary goal is to synthesise DNA with a free terminal end and polymerase initiation point. A pair of primers, one at the template strand and the other at the complementary strand, bind to opposing ends of the sequence being constructed; similarly, the 3' corresponds to the template strand for the elongation step.
The front primer takes 3'-5' and the reverse primer takes 5'-3'. However, the process of elongation culminates in the formation of two additional strands of ds DNA.
Caution: Dimer formation happens when the template strand is next to the complimentary strand, which is an undesirable circumstance.
Forward and reverse primers do not obey the complementarity rule; instead, a forward primer binds to one end of one target at 5' P, while the reverse primer occupies the opposite end of 5' P.
Unlike replication, DNA primers were most commonly utilised in PCR because:
- RNA primers cannot be removed after the reaction since it is a unidirectional method of polymerization.
- DNA primers are simple to make.
- However, because the bulk of the amplification is done using DNA, it is advised that only particular nucleotide primers be used.
- DNA Primers: Invitro: PCR amplification, DNA sequencing.
- RNA Primers: In vivo: DNA replication, cloning.
- DNA primers: The process of amplification is temperature dependent with fewer proteins.
- RNA primers: The replication process is a catalytic reaction in an enzyme-dependent manner with several proteins.
- DNA primers: 18- 24 base pairs.
- RNA primers: 10- 20 base pairs.
- DNA primers are chemically manufactured, whereas RNA primers need the Primase enzyme.
- Although DNA polymerase initiates the addition of nucleotides to the reactive 3' (OH) end of existing nucleic acid, along with the elongation and replication of the parent strand, RNA primers are shorter-lived and more reactive.
- Primer length: Oligonucleotides with a length of 18-24 are believed to be silent and beneficial, allowing short primers to quickly bind to the template at the annealing temperature.
- Melting point (52°C-56°C) The sequence's GC findings provide a good indication of the primer Tm. However, the primer difference should not be less than 2°C.
- Primer Annealing (Ta): A high Ta yields a low PCR product due to inadequate primer-template hybridization, whereas a low Ta yields non-specific PCR products due to a high number of base pair mismatches.
Ta = 0.3*Tm (primer) + 0.7 (product) – 14.9. (primer)
- Melting Point of the Primer:
Tm (primer)- It is used to calculate the least stable primer-template pair.Tm (product)- This parameter determines the melting temperature of the PCR product.Gradient PCR may be used to execute the modified step annealing, and the temperature can be tuned to bind primers.
- Clamp: Gene Sequencing and Primer GC Content Primers must have a GC concentration of 40-60%, with the 3' end containing two GC bases- GC clamp. However, GC bp with three H bonds is stronger than AT bp with two bonds due to the primer's high stability as well as the enhancement and specificity of the primer binding.
- Setting Restriction Enzyme (RE) Cut Sites: The enzyme known as the leader sequence allows for increased effectiveness for cutting enzymes by adding 3-5 bases to the 5' end of our target primer's overall cut site.
- End Stability: The greatest G permits the 3' end to bind 3-5 least bases. A sturdy 3' end, on the other hand, can help to prevent erroneous priming.
- Hairpins: Tolerable is the loop shape created by intramolecular interactions within the primer, which has an optimum 3' end with -2kcal/m and an internal hairpin with -3kcal/m.
- Dimers: A structure formed by intermolecular interactions between two primers to generate ds DNA. Similarly, interactions established between two homologous or the same sense of primer are referred to as self-dimers, but interactions created between opposing primers are referred to as cross dimers.
- Repeats & Run: The most essential characteristic is the continual occurrence of dinucleotide runs in a continuous stretch of a single nucleotide. The maximum number of repetitions and runs allowed was four dinucleotides and four base pairs.
- Primer- Template Cross Homology: Primers should be built in such a way that no homology within the template other than the target site is detected, resulting in non-specific binding and amplification. This may be divided into two categories:
a) Intra-primer homology: In the area of more than three bases, complimentary bases within the same pair can produce intramolecular bonding.b)Inter-primer homology: Intermolecular bonding is caused by forward and reverse primers with complementary sequences.
Primer design Protocol/ steps/Process
Golden Gate Cloning Method of Primer Designing
This approach necessitates the incorporation of a vector assembly (plasmid) into a single construct containing one or more DNA pieces. The PCR primers overlap to produce restriction sites with neighbouring DNA fragments and are designed for directed assembly using Type 2S enzyme and fragment DNA ligases. Similarly, this approach makes use of the type 2 class of restriction sites by cutting outside of their restriction sites via non-palindromic sticky end overhangs. This approach makes use of several DNA fragments by combining overhang sequences on their insert segments for facile annealing with the surrounding ones. Type 2S restriction sites, on the other hand, allow for golden gate assembly, leaving no restriction sites following cloning.
Primer Design using Gibson Cloning Method
Despite restriction digestion and ligation, this approach relies on recombination to generate plasmids. It enables the generation of identical homologous sequences 20-40 bp length on either side of the linearized vector for both target DNA pieces. This combines an identical overlap that is cleaved by exonuclease so that complementary parts can overhang it during assembly. In a nutshell, this is a strategy that evaluates two sections of homologous DNA combined with overlapping ends of DNA that fuse with them.
Validation of Primers and Probes in qPCR
Once the qPCR primers and probes have been designed using accessible tools, BLAST (insilico validation) will be used to check the specificity of the targeted gene sequences. The BLAST method searches for sequence similarity against various databases using a collection of gapped alignments of links to complete database entries (Raymaekers M et al, 2009). The query coverage and maximum identity should both be 100%. The BLAST software, on the other hand, reports a statistical significance, known as "expectation value" (E – value), for each alignment, which is an indicator for discovering the match by chance. E – values less than 0.01 represent homologous sequences (Altschul et al., 1990). ( Karlin et al., 1990). E-value metrics are used to evaluate putative biological linkages (Raymaekers M et al, 2009). Despite the fact that Insilco tools give useful feedback, the specificity of the qPCR assay employing the chosen primers and probes must be empirically confirmed using direct experimental data (Bustin et al., 2009).


~1.webp)

.webp)

.webp)
