The temperature-dependent nucleic acid amplification method known as polymerase chain reaction (PCR) is used to enzymatically amplify DNA or RNA in vitro.
It was created by Kary Mullis and his collaborators in the middle of the 1980s and is one of the most powerful and crucial tools in molecular biology and genetics today. The principles of nucleic acid replication and hybridization are combined in this process. This non-culture-based nucleic acid amplification method allows us to quickly create billions of copies of a single DNA or RNA segment.
Since its creation, PCR has undergone a number of changes, leading to the availability of many PCR procedures for various uses. The two PCR kinds that are most often employed are reverse transcriptase PCR and quantitative PCR (qPCR).
Reverse transcriptase polymerase chain reaction, or RT-PCR, is a specific kind of PCR method that in vitro amplifies RNA enzymatically.
It is the only kind of PCR that has the capacity to amplify RNA. The reverse transcriptase enzyme is used in addition to the other fundamental PCR building blocks.
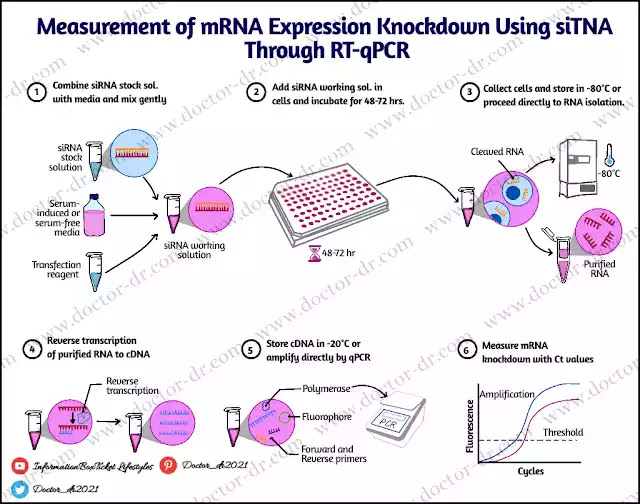
First, reverse transcription, which is mediated by the reverse transcriptase enzyme, converts the sample RNA to complementary DNA (cDNA). The PCR procedure uses these cDNA molecules as a template for amplification.
mRNA or micro RNA analysis and gene expression research are done using RT-PCR.
Table of Contents
- Requirements (Enzymes) of RT-PCR
1. Nucleic Acid Sample (Sample RNA)
2. Reverse Transcriptase Enzyme
4. Primers (Oligo (dT) primers, random primers, and sequence-specific primers)
5. Deoxynucleotide Triphosphates
6. PCR Buffers and Other Chemicals
Objectives of RT-PCR
- to make billions of copies of a single RNA segment by amplifying the particular RNA segment.
- to identify certain illnesses, genes, and to investigate gene expression.
Principle of RT-PCR
The reverse transcription and standard PCR processes are combined in RT-PCR. The reverse transcriptase enzyme initially converts the sample RNA to double-stranded DNA (complementary DNA) during the reverse transcription process. The two single-stranded DNA templates can then be created from the cDNA by heat decomposition. Based on the nucleic acid hybridization principle, primers can anneal to their complementary sequences in these ssDNA templates. Then, using the DNA replication principle, DNA polymerase lengthens the primer by successively adding the nucleotides to the 3' end to create a dsDNA. Denaturation, annealing, and elongation are three procedures that are cycled through repeatedly in order to produce millions of copies of the cDNA while controlling the temperature of the reaction.
Requirements (Enzymes) of RT-PCR
1. Nucleic Acid Sample (Sample RNA)
RT-PCR uses RNA as its sample, as opposed to other PCR methods that use DNA. The sample used most frequently is mRNA. Prior to amplifying, the RNA will be transformed into cDNA.
2. Reverse Transcriptase Enzyme
It is an enzyme that causes the RNA strand to split into two complementary DNA (cDNA) strands. It produces central-dogma reverse and goes by the name of RNA-dependent DNA polymerase enzyme. It is a crucial part of RT-PCR because it turns sample RNA into cDNA for amplifying.
3. DNA Polymerase Enzyme
By assembling the nucleotides sequentially in accordance with the template strand, DNA polymerases are enzymes that catalyse the synthesis of complementary DNA strands. The most extensively used DNA polymerase is Taq DNA polymerase, which was isolated from the bacterium Thermus aquaticus. Taq DNA polymerase is thermally stable and keeps working even after repeated cycles of heating and cooling.
4. Primers (Oligo (dT) primers, random primers, and sequence-specific primers)
In RT-PCR, three main kinds of primers are utilised;
1. Random Primers
For the purpose of cDNA synthesis using reverse transcriptase, these are the brief single-stranded sequences of 6–8 nucleotides that bind at the complementary site of RNAs with or without poly(A).
2. Oligo (dT) Primers
They are repeating deoxythymidine (dT)-containing oligonucleotides, typically 12–18 nucleotides long, that bind to the polyA tail of mRNA.
3. Sequence-specific Primers
These are the brief single-stranded nucleotide sequences that bind to the particular area of the sample RNA that is of interest. Most often, it is utilised in one-step RT-PCR.
5. Deoxynucleotide Triphosphates
Deoxynucleotide triphosphates (dNTPs) are synthesized nucleotides that are used in cDNA synthesis and the creation of additional strands of cDNA during amplification. Deoxyadenosine triphosphate (dATP), deoxyguanosine triphosphate (dGTP), deoxythymidine triphosphate (dTTP), and deoxycytidine triphosphate are the four distinct dNTPs that are employed (dCTP).
6. PCR Buffers and Other Chemicals
7. Thermocycler (PCR Machine)
Types of RT-PCR
RT-PCR may be divided into two forms depending on whether the reverse transcription and the amplification stages take place in one reaction (or tube) or two separate reactions (or tubes):
1. One-Step RT-PCR
It is a kind of RT-PCR in which the amplification and reverse transcription processes take place in the same tube. One tube is used to combine all the necessary ingredients. Reverse transcription first takes place, creating cDNA, which is later amplified by PCR.
Advantages of One–Step RT – PCR over Two–Step RT – PCR
- It features an easy setup and handling.
- It is more precise and accurate.
- It is less likely to be contaminated.
- It is a quicker and less expensive way.
Disadvantages of One-Step RT-PCR over Two-Step RT-PCR
- Due to the use of many chemicals in a single reaction tube, it detects fewer templates per reaction combination.
- It needs a bigger template to start since the template detection is less accurate.
- It prohibits the storage and subsequent examination of the cDNA produced during the reaction.
- Primer - dimer and non-specific binding have a higher likelihood.
- Reaction failure is a somewhat common occurrence.
2. Two-Step RT-PCR
It is a different kind of RT-PCR in which reverse transcription and amplification take place in two different tubes. Reverse transcription takes place in the first tube, producing cDNA. The PCR mixture is then introduced to another tube with the cDNAs there, where they are amplified.
Advantages of Two-Step RT – PCR over One-Step RT – PCR
- We can save cDNA produced by reverse transcription thanks to it.
- Greater efficiency, precision, and dependability are also present, and more templates may be detected per reaction mixture.
- Reduced likelihood of primer-dimer bonding, non-specific binding, and reaction failure.
Disadvantages of Two–Step RT – PCR over One-Step RT – PCR
- The likelihood of contamination is increased.
- It is a more difficult and laborious procedure that needs more resources and a skilled individual.
Steps/Procedure of RT-PCR
Reverse transcription and amplification are the two steps that make up the fundamental process. The process differs from one-step to two-step RT-PCR as well. However, the fundamental procedures are the same for both types and may be broken down into four stages: the preparatory stage, reverse transcription stage, amplification stage, and the product analysis stage, i.e.
1. Preparatory Stage
RNA extraction and the preparation of the whole reaction mixture are done at this initial stage. Prior to extracting the sample or bringing it from storage, all materials must first be organised, safety precautions followed, the PCR reaction preparation environment sanitised, and all reagents brought to working temperature.
In the one-step RT-PCR, Primer, DNA polymerase, DNA polyadenylase, dNTPs, buffers, reverse transcriptase enzyme, RNase H, and all other components are added in a predetermined and calculated amount to a single reaction tube. Following that, the tube is placed into a thermocycler for further processing.
In the two-step, RT-PCR, Reverse transcriptase, RNase H, primers, dNTPs, and other buffers and chemicals are added into a tube along with the sample RNA. The tube is then placed in a thermocycler and heated to a certain temperature, where cDNAs are produced.
2. Reverse Transcription
The RNA is first transformed into cDNA in this stage, which is followed by amplification.
In a tube, the entire reaction mixture—including reverse transcriptase, RNase H, dNTPs mixture, primers, nuclease-free water, reverse transcription buffer, and other components in one-step RT-PCR—is added. The tube is then heated in a thermocycler to a temperature of 40 to 50 °C for 10 to 30 minutes to complete the amplification process. The primer will attach to the appropriate RNA sample site at this temperature, and the reverse transcriptase enzyme will create cDNA by adding the free dNTPs.
3. Amplification
4. Product Analysis Stage
Applications of RT-PCR
1. Study Gene Expression
2. Identification of Unknown Species
3. Infectious Disease Diagnosis
4. Gene Insertion and Gene Therapy Study
5. Study Mutation and Cancer Cells
6. Tools of Genetic Engineering and Viral Study
Advantages of RT-PCR
- Enzymatically, millions of copies of mRNA may be produced using this quick process of RNA amplification.
- It is quite easy to use. A thermocycler controls and operates the process semi-automatically without the need for human intervention.
- It is affordable yet has a very high specificity and sensitivity.
- It is a fairly precise approach for determining whether or not an individual has been exposed to an RNA virus. RNA viruses can be divided into strain-level classifications. It has reduced the time needed to detect viral infections and RNA viruses.
- Compared to the conventional Northern Blot method, it can only identify a very little quantity of mRNA (around 5pg).
- Gene expression and mutated genes may be quickly and readily investigated. This has made it feasible to research gene insertion, identify cancer in its early stages, and track the effectiveness of gene therapy.
- It may be used to both identify and measure the sample RNA because it is a qualitative and quantitative procedure.
Limitations of RT-PCR
- Only RNAs, especially mRNAs, may be amplified by it.
- For the purpose of creating primers, prior knowledge of the RNA sequence is necessary.
- Because the system depends on enzymes and operates at full temperature, even a little variation in the reaction temperature will reduce the enzyme's effectiveness. Therefore, a stringent system of temperature regulation is needed.
- A little amount of contamination with a comparable primer binding site can be magnified and provide an incorrect result, either positive or negative.
- A tiny quantity of organic or inorganic contamination in the reaction mixture can have a significant impact on the reaction.
- It takes a complicated reaction mixture and a trained individual to run the procedure, which is quite laborious.



~1.webp)
.webp)
.webp)

