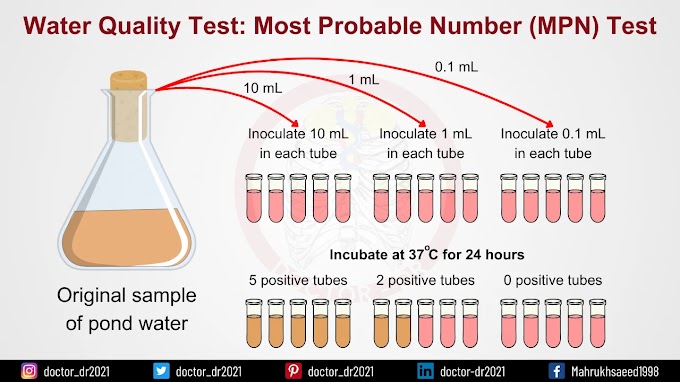
Introduction
Welcome to our in-depth exploration of microbiology techniques and tools for whole genome sequencing of bacterial genomes. In this educational journey, we will uncover groundbreaking insights into the fascinating realm of microbes. Specifically, we will focus on the powerful analysis techniques used for species identification, multilocus sequence typing (MLST), and detecting resistance genes in bacterial genomes.
Kmer Finder: Harnessing Whole Genome Sequencing Data for Species Identification
When encountering bacterial organisms in our research, a fundamental question arises: what species is it? Traditionally, the 16s rRNA gene has been pivotal for sequence-based taxonomy. However, alternative methods, such as utilizing the GB Gene ribosomal MLST or specific protein domain profile Fes, have been proposed to overcome its limitations.
Whole genome sequencing offers comprehensive genetic insights by harnessing the entirety of genetic data encoded in the entire genome. This is where Kmer Finder comes into play. Kmer Finder utilizes short DNA sequences called "kers" to infer species from whole genome sequencing data. By comparing these kers across different organisms in a database, Kmer Finder can pinpoint the best matching species.
The efficiency of Kmer Finder is optimized through the minimization of redundant data in the database. By retaining only specific kers with particular prefixes, the tool ensures swift and efficient species prediction performance. Extensive testing has shown that Kmer Finder outperforms existing tools in predicting both genus and species levels with superior accuracy.
Result Interpretation
When analyzing the results from Kmer Finder, it is important to focus on the best matching species and consider parameters such as kmer score, template length, template coverage, query coverage, and depth. These metrics provide valuable insights into the reliability of the prediction. For example, a high kmer score, significant template coverage, substantial query coverage, and depth close to 1 indicate a strong match between the query and reference genomes, thus suggesting a high confidence in identifying the species.
Multilocus Sequence Typing (MLST): Characterizing Bacterial Strains
After species identification using Kmer Finder, multilocus sequence typing (MLST) is used to further characterize and classify bacterial strains based on the sequences of specific genes. MLST offers a higher resolution compared to kmer analysis alone by focusing on conserved housekeeping genes across bacterial genomes.
MLST targets a set of universally present housekeeping genes across bacterial species, ensuring consistency and comparability of results across different studies and laboratories. It provides higher discriminatory power compared to kmer analysis alone, as it can differentiate closely related strains that might have identical or very similar kmer profiles.
MLST is widely used in epidemiological studies to track the spread of bacterial strains, understand outbreaks, and study the population structure of bacterial pathogens. The sequence data generated through MLST can be easily shared and compared between different research groups and public health agencies.
MLST data also has clinical implications, such as identifying antibiotic resistance patterns or predicting virulence potential. Understanding the genetic makeup of bacterial strains through MLST can help guide treatment strategies and inform public health interventions.
Resfinder: Identifying Acquired Antibiotic Resistance Genes
Resfinder is a tool specifically designed to identify acquired antibiotic resistance genes within bacterial genomes or plasmids. These resistance genes confer resistance to various antibiotics and are crucial for understanding antibiotic resistance profiles and patterns.
The results from Resfinder have important clinical and epidemiological implications. They provide insights into the potential antibiotic resistance profile of bacterial strains, which is critical for guiding antibiotic treatment strategies in clinical settings and for monitoring the spread of antibiotic resistance in the community and healthcare settings. Resfinder data can inform public health interventions aimed at controlling the spread of antibiotic-resistant bacteria.
Resfinder results are also valuable for research purposes, such as studying the mechanisms of antibiotic resistance, tracking the evolution of resistance genes, and investigating the genetic basis of antibiotic resistance in bacterial pathogens.
Plasmid Finder: Studying Mobile Genetic Elements
Plasmid Finder is a tool frequently used in bacterial genomics and epidemiology to study mobile genetic elements like plasmids. Plasmids are extrachromosomal genetic elements that can carry a variety of genes, including antibiotic resistance genes. By identifying and characterizing plasmids within bacterial genomes, researchers can gain insights into the dynamics of horizontal gene transfer and the dissemination of antibiotic resistance genes within bacterial populations.
Plasmid Finder helps in understanding the prevalence, diversity, and distribution of plasmids among bacterial strains. By correlating Plasmid Finder results with the antibiotic resistance profile obtained from Resfinder, researchers can investigate the association between plasmids and antibiotic resistance. This information is crucial for understanding the mechanisms underlying the spread of antibiotic resistance and for developing strategies to combat it.
Plasmid Finder data is also valuable for various research purposes, including studying the evolution of plasmids, tracking the spread of plasmid-mediated antibiotic resistance, and investigating the role of plasmids in the adaptation and survival of bacterial pathogens in different environments.
CSI Phylogeny: Constructing Phylogenetic Trees
CSI Phylogeny is a tool used to construct phylogenetic trees based on whole genome sequence data of bacterial isolates. By identifying core genomic regions shared among the isolates, CSI Phylogeny ensures that the phylogenetic analysis is based on stable and reliable genetic information.
The resulting phylogenetic tree generated by CSI Phylogeny provides a visual representation of the evolutionary relationships among the bacterial isolates. It depicts how closely related or distant the isolates are from each other based on their genetic similarity.
Researchers can interpret the phylogenetic tree generated by CSI Phylogeny to understand various aspects of strain relatedness, such as the genetic diversity within a population, the evolutionary history of the isolates, and potential transmission pathways in epidemiological investigations.
Practical Assignment: Identifying Outbreak Strains
Now, let's put our knowledge into practice. You will be given a file containing five unknown samples and an Excel file to summarize your results. The objective is to identify three strains that are outbreak strains.
You will start by using Kmer Finder to determine the species of each unknown sample. By comparing the kmer scores, template length, template coverage, query coverage, and depth, you can identify the best matching species for each sample.
Next, you will use MLST to characterize the bacterial strains further. MLST typing will provide you with sequence types for each strain, helping to differentiate closely related strains.
After MLST, you will use Resfinder to identify acquired antibiotic resistance genes within the genomes or plasmids of the characterized bacterial strains. This will give you insights into the antibiotic resistance profiles of the strains, which is crucial for understanding their pathogenic potential.
Following Resfinder, you will employ Plasmid Finder to identify and characterize plasmids within the bacterial genomes. This step will help you understand the role of plasmids in the spread of antibiotic resistance among bacterial strains.
Finally, you will use CSI Phylogeny to construct a phylogenetic tree based on the whole genome sequence data of the bacterial isolates. The tree will illustrate the evolutionary relationships among the isolates and help you determine the most distantly related strain.
By analyzing the results from all these tools, you will be able to identify the outbreak strains based on species, MLST typing, antibiotic resistance profiles, and phylogenetic relationships.
Conclusion
In summary, the tools and techniques discussed in this blog post empower researchers to accurately characterize bacterial species, track their evolutionary paths, and uncover crucial insights into antibiotic resistance. Kmer Finder harnesses the power of whole genome sequencing data and kmer analysis for species identification. MLST provides a standardized method for strain typing, epidemiology, and evolutionary analysis. Resfinder identifies acquired antibiotic resistance genes, and Plasmid Finder studies plasmids' role in the spread of antibiotic resistance. CSI Phylogeny constructs phylogenetic trees to understand strain relatedness. By combining these tools, researchers can gain a comprehensive understanding of bacterial populations and their dynamics.
We hope this blog post has provided you with valuable insights into the world of microbiology techniques and tools for whole genome sequencing. By leveraging these powerful analysis techniques, researchers can continue to make groundbreaking discoveries and contribute to the field of microbiology.